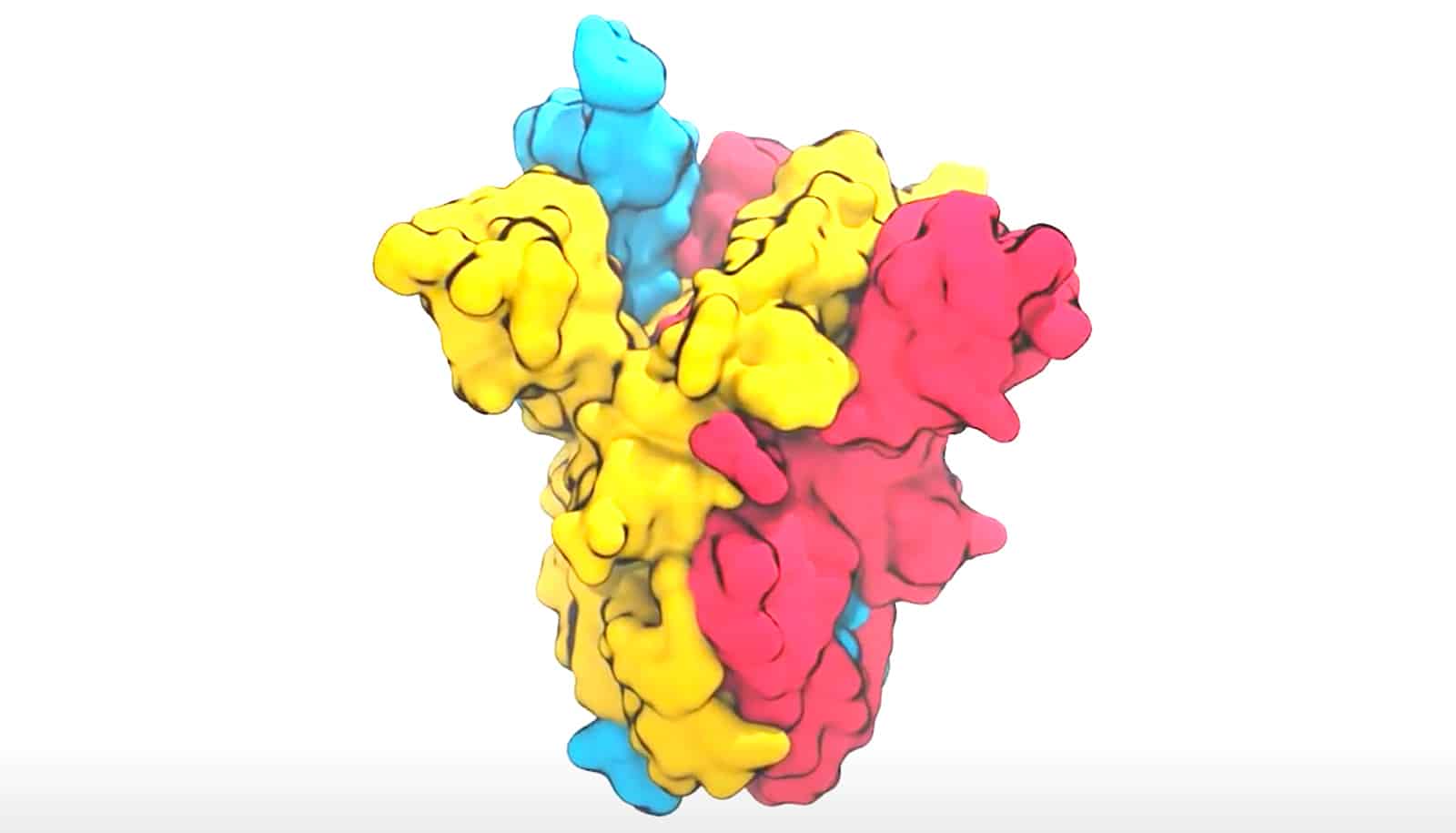
New research on a “spike” protein will sift through 1 billion drug-like molecules and 60 sites on the new coronavirus to find most promising options for targeted drug development.
To create computer models to better understand how the “spike” protein on the surface of the COVID-19 virus interacts with the cells it infects, a team of Stony Brook University researchers is working with scientists at the US Department of Energy’s (DOE) Brookhaven National Laboratory and Argonne National Laboratory, and will be leveraging those laboratories’ computational resources and expertise.
These spikes all around the exterior of the spherical virus particles give the virus the look of a crown and its name as a coronavirus. They are also the part of the virus that attaches to a cell inside people’s respiratory systems, using a specific protein on the human cell to begin the process of causing infection. Because of this role, the spike protein is also the main target for antibodies that would provide immunity to coronavirus and for developing drugs that potentially could block infection.
Scientists have already captured useful “pictures” of the spike with atomic-level detail, and they have a good idea of what the spike is doing. For example, they know that the spike changes its shape as it contacts the cell, and this leads to the virus and cell merging. But they need to know more detail in order to control it.
“You may know that your car door is the way you can get in and out of a car, but if you’ve only seen the door in a picture and have never watched someone actually open a door, you don’t really know how it works,” says team leader Carlos Simmerling, professor of physical and quantitative biology, a professor of chemistry, and associate director of the Laufer Center at Stony Brook University.
“You also probably would not guess that the handle is the part that you need to move to get the door to open, and that perhaps there is a key that can lock it and keep it from opening.
“With our research and computer modeling, we are working to understand the process at that more detailed level,” he explains. “For example, what exactly leads to the spike of the virus opening the ‘door’ to infection when it contacts the cell? And, are there places on the spike away from this contact point that could act like keys, where scientists might be able to make a small molecule that would bind to it and lock it closed?”
As Kerstin Kleese van Dam, director of Brookhaven Lab’s Computational Science Initiative, notes, “We will be looking at 60 different target sites where a new drug may attach to the virus and 1 billion drug-like molecules that might be used to modify the ‘door handles,’ ‘hinges,’ or other components so we can identify the most promising options for neutralizing the virus to keep it from entering cells.”
This work is taking place with funding from Brookhaven Lab’s program development funds, Stony Brook University, and Argonne National Laboratory’s Computing, Environment, and Life Sciences Directorate through DOE’s Exascale Computing Project and the National Institutes of Health.
Source: Stony Brook University