Researchers have determined the atomic interactions that stabilize the liquid yet “condensed” phase of FUS, an important phase-changing protein linked to severe cases of amyotrophic lateral sclerosis and a certain type of dementia.
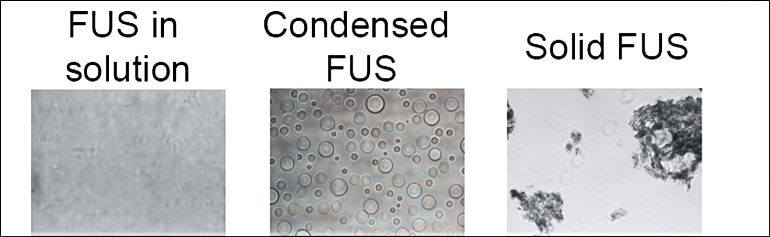
Most of the well-studied proteins in our bodies are like metal; some can change shape easily, like aluminum foil, and others are rigid, like steel beams, but they typically have a solid, well-defined structure. Many other essential proteins are more like water—able to change phase from liquid to solid ice.
FUS is one of these important phase-changing proteins. In healthy cells, FUS switches between floating diffusely and condensing into liquid droplets with other proteins to make, edit and deliver the blueprints for protein production. However, FUS also has a “solid” or aggregate phase that has been found in some people with severe cases of amyotrophic lateral sclerosis (ALS) and a type of dementia called frontotemporal dementia.
“What we want to understand is the atomic details of these interactions so that we know what kind of treatments would be important for ALS and other diseases,” says co-corresponding author Nicolas Fawzi, an associate professor in the molecular pharmacology, physiology, and biotechnology department at Brown University.
“First we need to know the structural differences between the normal form and the disease form, so we know where to put in a wrench to stop it. We can’t design a drug to bind to something when we don’t know what it looks like.”
FUS protein droplets
Fawzi says you can think of the condensed liquid droplets FUS can form within cells as somewhat like the condensation that forms on a cold glass on a humid day. Both the water droplets and the humid air contain water molecules, but they are in distinct phases.
In addition to its implication in ALS—commonly called Lou Gehrig’s disease—the disordered region of FUS is known to be associated with some types of cancer including Ewing’s sarcoma, Fawzi says. In fact, the acronym FUS stands for FUsed in Sarcoma.
Using a combination of nuclear magnetic resonance (NMR) spectroscopy, Raman spectroscopy, and computational modeling, the researchers found that the interactions between a disordered region of FUS in the condensed liquid droplets are quite varied and dynamic, he says. FUS does not form any traditional structural elements.
However, by combining experiments with computational modeling, the team demonstrated that multiple atomic interactions—including those formed by specific amino acids within the protein, namely glutamine and tyrosine—maintain the condensed yet disordered nature of FUS.
Guiding future treatments?
The knowledge of these molecular interactions—which are distinct from the interactions in the neurodegeneration-associated aggregate phase—might someday be used to guide the development of therapeutics that hinder disease-associated aggregation or support the normal condensed phase interactions.
Additionally, other scientists could use the combination of techniques Fawzi’s team used to uncover FUS’s atomic interactions to study intrinsically disordered proteins that also form liquid or solid condensed forms, such as Huntington’s, Parkinson’s, prion diseases, and Type II diabetes in addition to ALS.
“We did these NMR experiments in a new way that allowed us to explicitly look at which atoms in one protein are interacting with atoms in another FUS protein,” Fawzi says. “I expect that people will start doing experiments like this because it provides more assurance and detail on the contacts between intrinsically disordered proteins. Disordered proteins do all kinds of important things, and we don’t really know how they normally work. When they go wrong, we really don’t know what’s going on.”
Fawzi plans to continue studying FUS. Specifically, he wants to study the entire protein, not just the intrinsically disordered region that was the primary focus of this paper, in test tubes as well as in living cells. He also plans to apply this combination of techniques to continue studying the molecular interactions of other neurodegeneration-associated disordered proteins such as hnRNPA2 and TDP-43.
The findings appear in Nature Structural and Molecular Biology.
Additional coauthors came from Lehigh University; Brown University, and the University of Texas at Austin. The National Institutes of Health, the National Science Foundation, the Human Frontier Science Program, the US Department of Energy, and Deutsche Forschungsgemeinschaft supported the research.
Source: Brown University



